Literature Sharing: Multiplex Immunofluorescence for MCC Tumor Microenvironment Research
Background
Merkel cell carcinoma (MCC) is an aggressive cutaneous neuroendocrine carcinoma that primarily affects older adults. The incidence is higher and occurs earlier in immunocompromised individuals, with a high rate of recurrence, metastasis, and mortality, necessitating effective prognostic evaluation and treatment approaches. MCC develops through two distinct pathways: most tumors are driven by clonal integration of the Merkel cell polyomavirus (MCPyV) and expression of oncogenic viral T antigens; MCPyV-negative tumors arise due to ultraviolet-induced high tumor mutation burden. Traditional predictors of immunotherapy response (such as PD-L1 expression and tumor mutation burden) are ineffective for MCC, while detailed analysis of the tumor microenvironment (TME) may more effectively predict treatment response. In-depth understanding of MCC-associated TME may provide new alternative treatment strategies for patients who are ineffective or intolerant to immunotherapy.
Transplanting mouse-derived tumor cells into mice with the same genetic background allows precise observation of the effects of immunomodulators on tumors, making it an important tool for anti-tumor immunity research. Therefore, the study "Development of a Multiplex Immunofluorescence Assay for Tumor Microenvironment Studies of Human and Murine Merkel Cell Carcinoma" by Sun L et al. utilized multiplex immunofluorescence (mIF) technology to simultaneously visualize multiple markers on a single tissue section, developing a standardized mIF detection method suitable for studying the TME of human and mouse MCC tumors.
Research Methods
First, ethically approved surplus human MCC tissues and mouse MCC samples following animal experimental guidelines were collected, all processed into formalin-fixed paraffin-embedded (FFPE) sections. Then, with the key MCC transcription factor SOX2 as the core tumor marker, mIF experiments were designed with T cells, macrophages, and species-specific functional markers. The mIF experiments were optimized through conditions such as primary antibody concentration, antigen retrieval conditions, staining order, and fluorophore selection. Manual staining was performed using the Opal 6-Plex kit, followed by imaging on the Mantra/Polaris platform and analysis with InForm software. Finally, the accuracy of cell population identification was verified by comparing with corresponding singleplex IHC results and phenotypic analysis, and the specificity of fluorescence signals and rationality of spatial distribution were evaluated.
Primary Antibody Localization Detection (Monoclonal Antibody Staining)

Although microwave treatment can be used in mIF detection to elute primary and secondary antibodies, avoiding cross-reactivity or signal residue, some epitopes may degrade or even disappear after multiple microwave treatments, while others may have enhanced signals. Therefore, the staining order cannot be set arbitrarily, and the optimal position for each antibody needs to be predetermined. The experiment used positive sections expressing target markers, with test primary antibodies arranged in different staining positions in the experimental group and primary antibody dilution buffer added to the primary antibody positions in the control group. Both groups underwent the same microwave treatment and replaced fluorophores with fluorophore dilution buffer. Then, the fluorescence intensity at each position was evaluated by fluorescence microscopy. The data in the figure clearly shows the signal performance of markers at different staining positions, where the position with positive cell signal intensity at least 5-10 times the background was determined as the optimal staining position.
Mouse mIF Marker Selection and Staining Verification
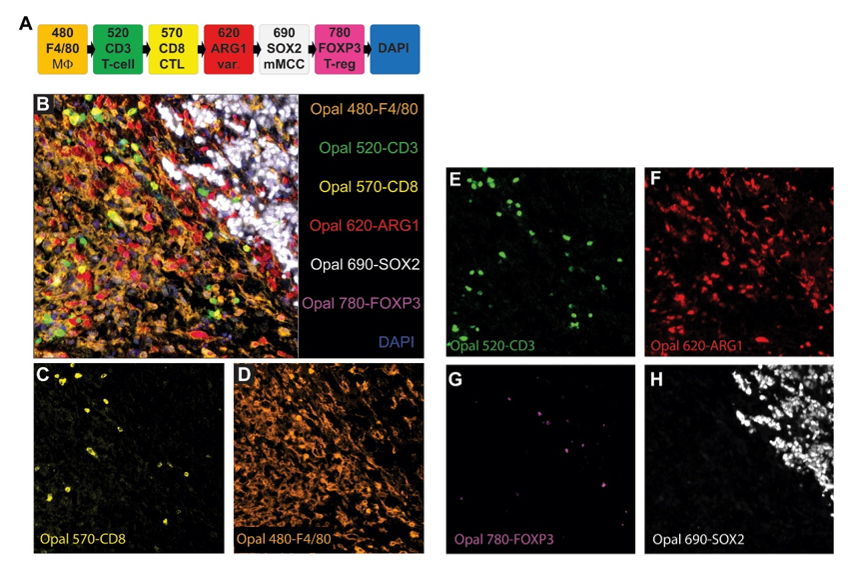
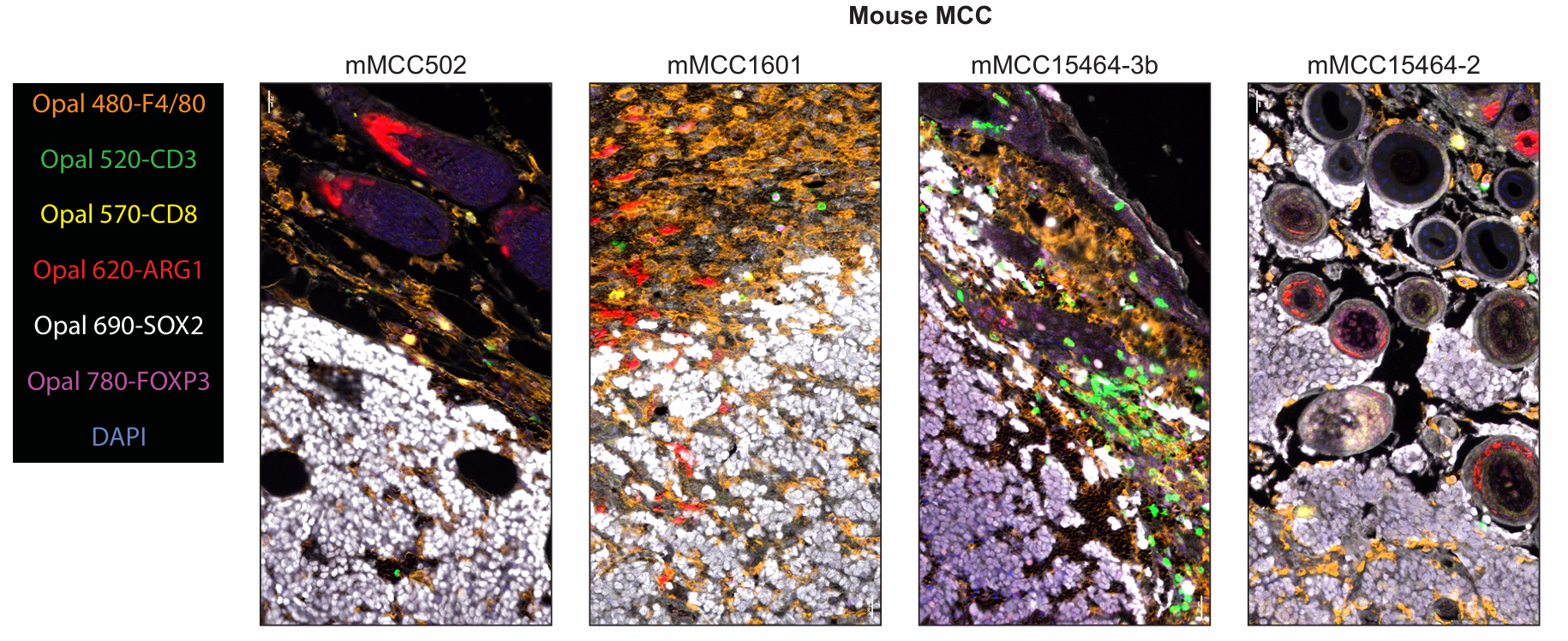
The figure shows the fluorophore matching and application effects of markers in mouse MCC tumor samples. SOX2 was paired with the lower-brightness fluorophore Opal 690 due to its high expression abundance; the signal intensity and staining position performance of FOXP3 supported pairing with Opal 780; the macrophage marker F4/80 was paired with Opal 480 despite high autofluorescence in the corresponding channel, as its signal performance was stable; Arg1 was paired with Opal 620 to maximize the staining position distance from F4/80; CD3 and CD8 were paired with Opal 520 and Opal 570, respectively. The figure clearly shows the multiplex staining images and single-channel images in mouse MCC samples. Meanwhile, the multiplex staining results of 4 additional mouse MCC tumor samples were similar to these, and the multiplex staining results were consistent with the singleplex IHC results of each marker, with phenotypic analysis accurately detecting the expected cell populations.
Human mIF Marker Selection and Staining Verification
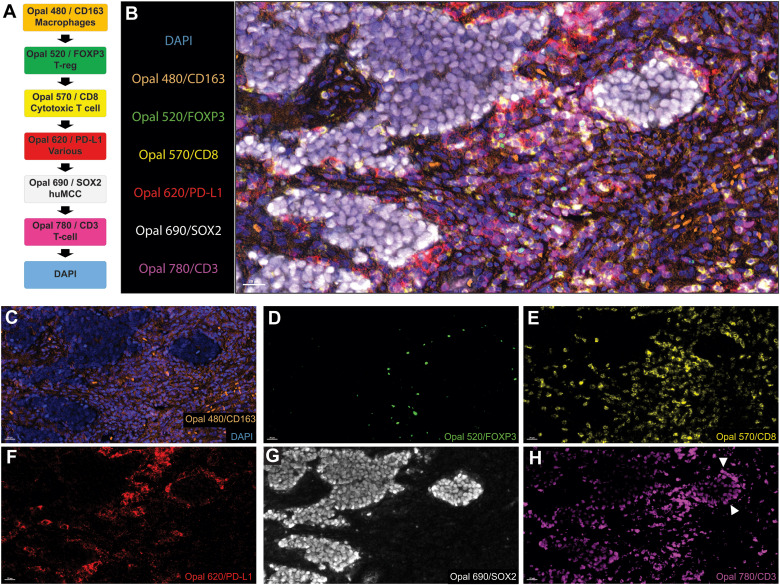
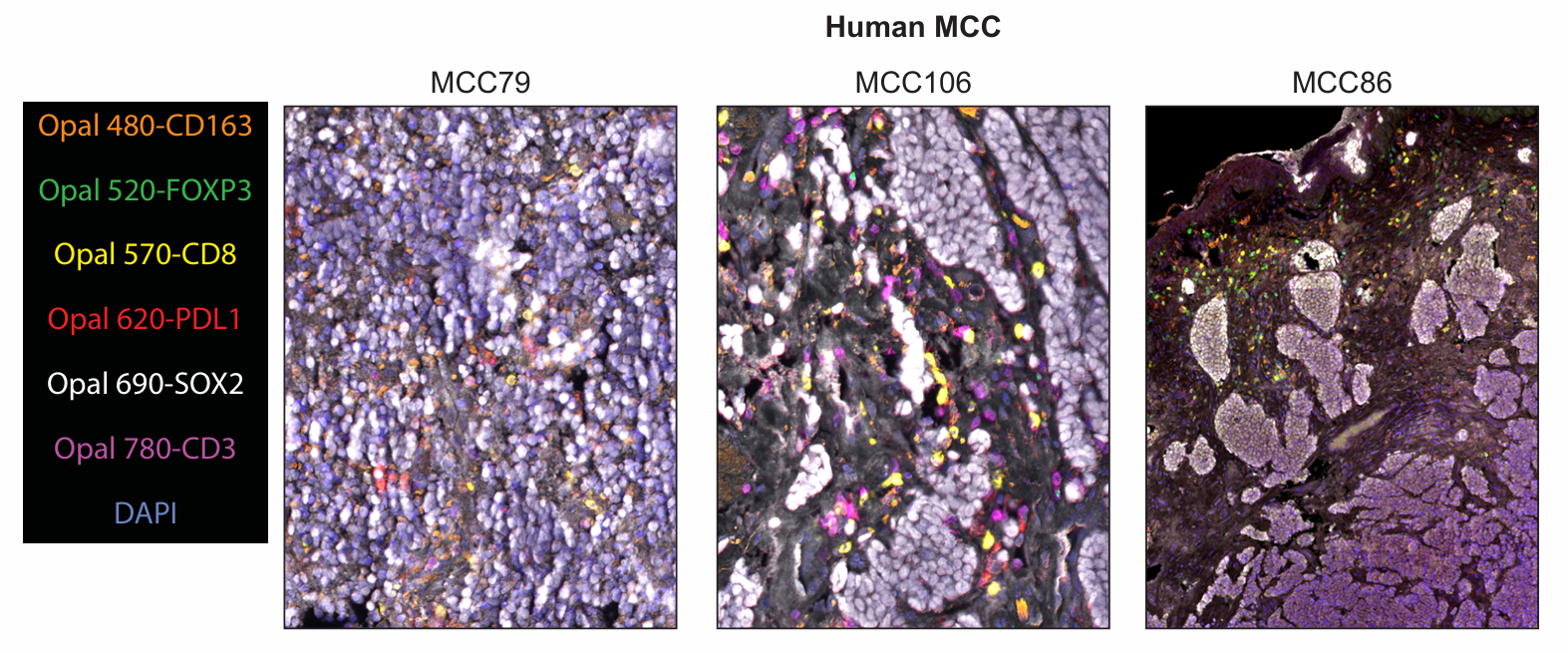
The figure shows the fluorophore matching and application effects of markers in human MCC tumor samples. Unlike mouse samples, CD3 and SOX2 as high-abundance antigens were paired with weak fluorophores Opal 780 and Opal 690, respectively; FOXP3 with weak expression was paired with the bright fluorophore Opal 520; the macrophage marker CD163 was predicted to be stably expressed, so it was paired with Opal 480 to tolerate autofluorescence interference in tissues; PD-L1 was paired with Opal 620, and CD8 was paired with Opal 570, to achieve staining position separation from CD163 and CD3, respectively. Although CD3 was located after CD8 in the final sequence, which theoretically had potential spatial hindrance risk, phenotypic analysis showed that the vast majority of CD8+ cells co-expressed CD3+ without classification errors. The figure clearly shows the multiplex staining images and single-channel images in human MCC samples, and staining results applied to 3 additional samples were similar to these.
Result Analysis Using InForm
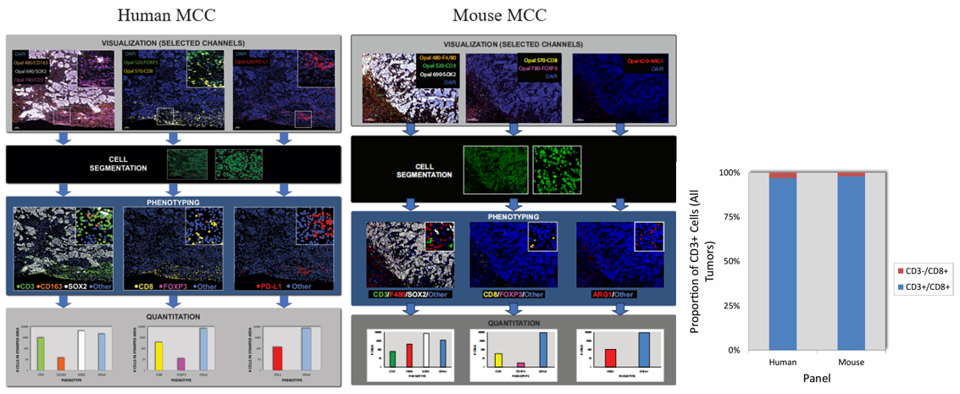
The analysis process is divided into four steps: the first step is to view each staining channel individually, in specific combinations, and as a whole; the second step is cell segmentation, defining regions in the image that represent individual cells; the third step is phenotypic classification, interpreting marker expression patterns through user-trained algorithms to assign phenotypes to each cell, with colored dots representing cells of different phenotypes; the fourth step is quantitative analysis, counting the number of each cell population. Results show that despite weak background labeling of tumor cells in the Opal 780 channel, SOX2+ tumor cells can still be correctly classified through the above digital phenotypic analysis, effectively distinguishing true CD3 expression signals on T cells from non-specific false signals on tumor cells. The staining order of CD3 and CD8 in both human and mouse tumor tissues is close, theoretically with spatial hindrance risk, but subsequent multiplex marker CD3+/CD8+ co-expression detection completed in R software did not significantly detect CD3-/CD8+ cells, indicating no false detection results due to spatial hindrance, ensuring detection accuracy. Moreover, mIF sections of both tumor tissues are suitable for this analysis method.
Conclusion
This study addresses the clinical and research pain points of insufficient prediction of immunotherapy response in Merkel cell carcinoma and lack of suitable TME detection tools, focusing on multiplex immunofluorescence technology, successfully developing and optimizing standardized mIF detection procedures suitable for human and mouse MCC. The study takes the highly specific MCC tumor marker SOX2 as the core, systematically optimizing key links such as antibody concentration, antigen retrieval conditions, staining order, and fluorophore matching. Verification through a series of result figures shows that both human and mouse MCC staining systems can accurately visualize target cell populations, with high phenotypic analysis accuracy, effectively distinguishing true signals from non-specific backgrounds, no obvious spatial hindrance, and good repeatability.
References
Sun L, Verhaegen ME, McGue J, Olivei AC, Dlugosz AA, Frankel TL, Harms PW. Development of a Multiplex Immunofluorescence Assay for Tumor Microenvironment Studies of Human and Murine Merkel Cell Carcinoma. Lab Invest. 2024 Oct;104(10):102128. doi: 10.1016/j.labinv.2024.102128. Epub 2024 Aug 23. PMID: 39182611; PMCID: PMC11502254.
Enkilife mIHC TSA Kits
Product | Catalog Number |
|---|---|
TSA Six-Label Seven-Color Multiplex Immunohistochemistry Kit | |
TSA Five-Label Six-Color Multiplex Immunohistochemistry Kit | |
TSA Four-Label Five-Color Multiplex Immunohistochemistry Kit | |
TSA Three-Label Four-Color Multiplex Immunohistochemistry Kit | |
TSA Two-Label Three-Color Multiplex Immunohistochemistry Kit |
