Lysine acetylation is a reversible post-translational modification that plays a crucial role in regulating protein function, chromatin structure, and gene expression[1]. Many transcriptional coactivators possess intrinsic acetyltransferase activity, while transcriptional corepressors are associated with deacetylase activity[2]. In response to signaling pathways, acetylation complexes (such as CBP/p300 and PCAF) or deacetylation complexes (such as Sin3, NuRD, NCoR, and SMRT) assemble to bind to DNA-bound transcription factors (TFs).
Histone acetylation by histone acetyltransferase (HAT) is associated with transcriptional activation, while histone deacetylation by histone deacetylase (HDAC) is related to transcriptional repression[3]. Histone acetylation promotes transcription by remodeling higher-order chromatin structure, reducing the interaction between histones and DNA, and providing binding sites for transcriptional activation complexes, which include bromodomain-containing proteins that can bind to acetylated lysinHistone deacetylation inhibits transcription through opposite mechanisms, including the assembly of higher-order chromatin structures and the exclusion of transcriptional activation complexes containing bromo domains[4]. Low histone acetylation is a hallmark of heterochromatin silencing. Increasingly, site-specific acetylation of non-histone proteins has been shown to regulate their activity,localization, specific interactions, and stability/degradation.
Notably, recent advances in mass spectrometry techniques have enabled high-resolution mapping of most acetylation sites in the proteome. These studies reveal that the 'acetylome' includes nearly 3600 acetylation sites in approximately 1750 proteins, indicating that acetylation modification is one of the most abundant chemical modifications in nature. In fact, this mark may affect the activity of proteins in various physiological processes, including chromatin remodeling, cell cycle, splicing, nuclear transport, mitochondrial biology, and actin nucleation. In terms of organisms, acetylation plays crucial roles in immunity, circadian rhythms, and memory formation. Protein acetylation is a promising target for the design of new drugs for many diseases[5,6].
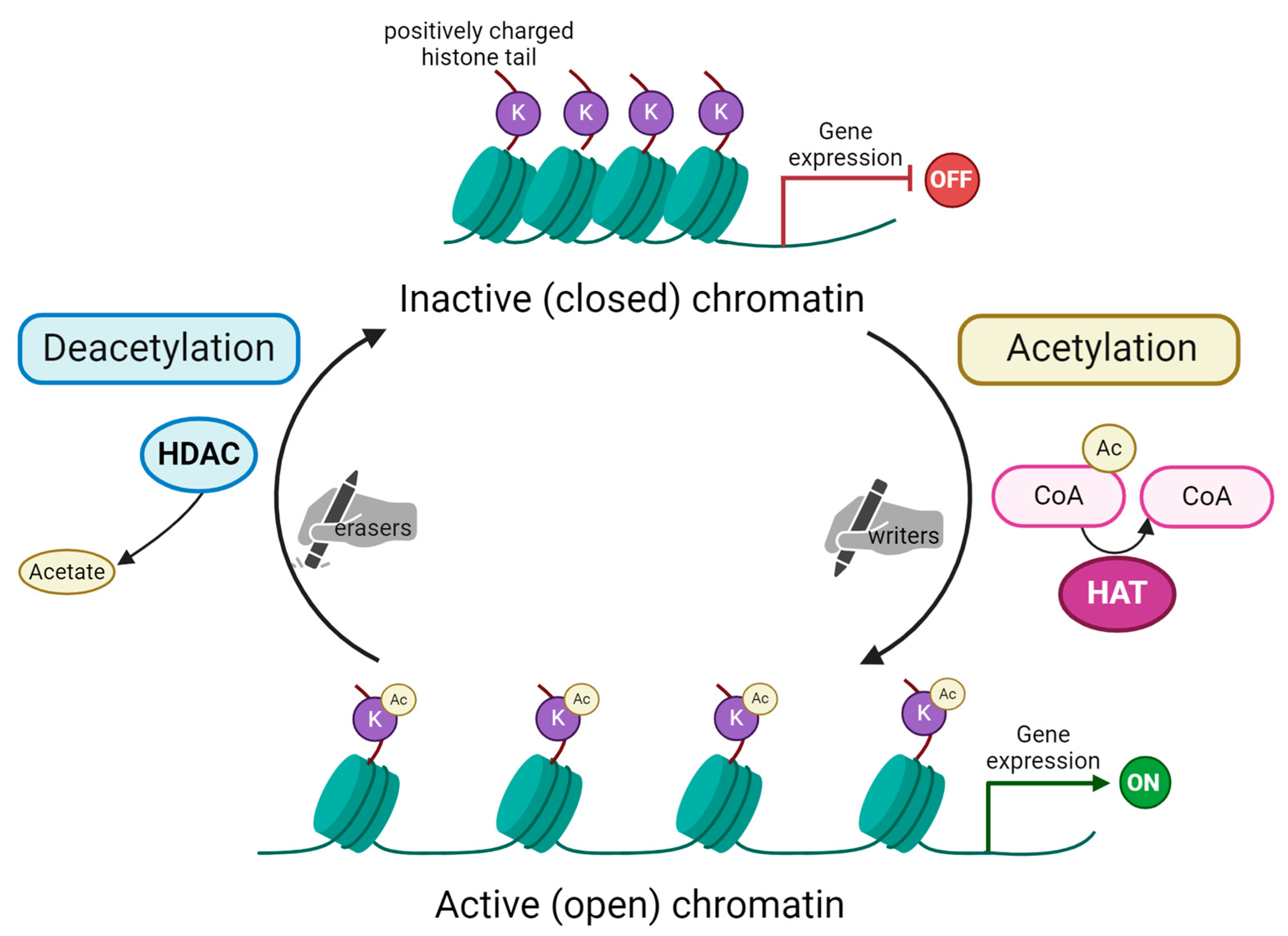
Schematic representation of histone acetylation and deacetylation process. Acetylation of lysine (K) residues on substrate histone protein is carried out by histone acetyltransferases (HAT). These enzymes are the so-called “writers” able to add acetyl groups mostly on histone tails. In the acetylation reaction, acetyl coenzyme A (here Ac-CoA) is converted into coenzyme A (CoA). Subsequently, acetylated histones impact chromatin’s structure, making it open and, as a consequence, activating gene expression. The process is reversible due to the action of histone deacetylases (HDAC), the “erasers” that remove the acetyl group from histone tails. This reaction releases acetate as a by-product. Chromatin remains in its condensed, inactive (closed) form. This blocks transcription machinery from accessing genes’ promoter regions and, in turn, represses gene expression. Created with
Relevant antibodies
| Catalog# | Product Name | Reactivity | Application |
|---|---|---|---|
| AMRe21336 | Histone H3 Rabbit Monoclonal antibody | Human,Mouse,Rat | WB,IHC,IF,IP,ELISA |
| APRab00847 | Acetyl-Histone H3 (Lys9) Rabbit Polyclonal Antibody | Human,Mouse,Rat | WB |
| AMRe03273 | Acetyl-Histone H3 (Lys14) Rabbit Monoclonal Antibody | Human,Rat | WB,ICC/IF,IP |
| AMRe84527 | Acetyl-Histone H3(Lys18) Rabbit Monoclonal Antibody | Human,Mouse,Rat | WB,IP,IHC,ICC,FC,IF |
| AMRe04172 | Acetyl-Histone H3 (Lys27) (17F16) Rabbit Monoclonal Antibody | Human,Mouse,Rat | WB,IHC-P,ICC/IF,FC |
| APS0635 | HRP-conjugated Polyclonal Goat Anti-Rabbit IgG(H+L) Secondary Antibody | Rabbit | ELIS, WB, Dot blot |
| AMre80004 | GAPDH (12R9) Rabbit Monoclonal Antibody | Human,Mouse,Rat,Rabbit,Dog,Monkey | WB,ELISA |
Related Products

