Key Strategies to Overcome Protein Inclusion Bodies in Escherichia coli Expression Systems
The Escherichia coli expression system is a cornerstone in recombinant protein production, favored for its well-characterized metabolic pathways, rapid cell growth, high protein expression levels, and clear genetic background. Despite these advantages, a significant challenge in using this system is the propensity for protein aggregation into inclusion bodies. These insoluble aggregates often lack biological activity, posing a considerable obstacle for researchers aiming to obtain functional proteins. Herein, we present five essential strategies to enhance the solubility of recombinant proteins in E. coli and mitigate the formation of inclusion bodies.
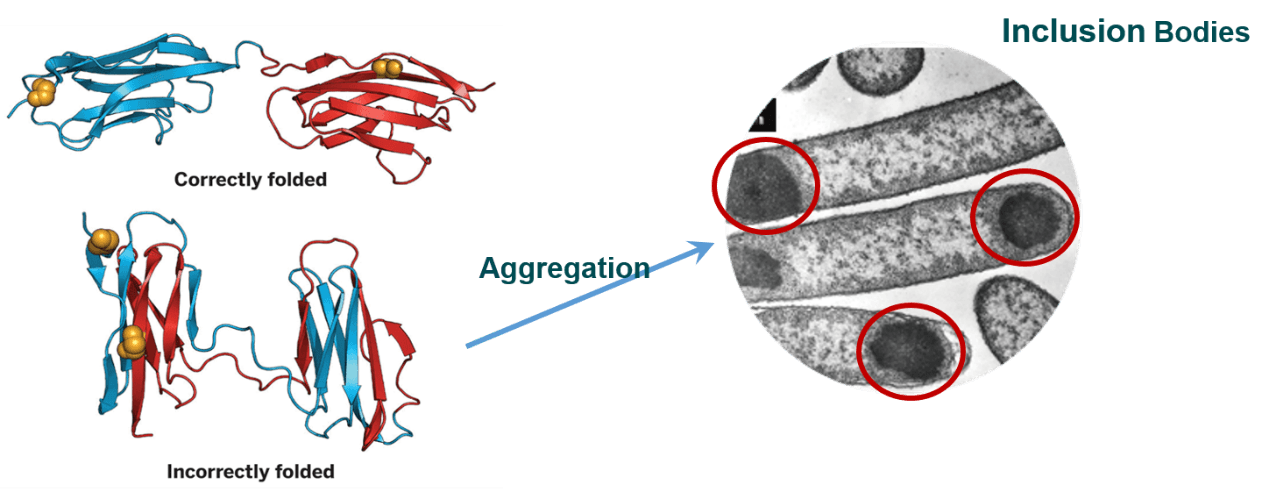
1. Removal of Signal Peptides
Signal peptides, typically consisting of 5-30 amino acids, play a crucial role in guiding newly synthesized proteins through secretory pathways and facilitating their folding into specific three-dimensional structures. They also determine the final localization of proteins to specific subcellular compartments. In the context of intracellular protein expression in E. coli, signal peptides are often functionally redundant and may even be detrimental. The hydrophobic nature of signal peptides can induce protein aggregation and the formation of inclusion bodies. Therefore, removing signal peptides from the target protein sequence is often recommended to promote soluble expression in E. coli (Baneyx, 1999).
2. Codon Optimization
During protein synthesis, tRNAs with corresponding codons guide amino acids to the mRNA for translation. In prokaryotic systems, the differential abundance of tRNAs leads to codon bias. E. coli contains eight rare codons: AGA, AGG, CGG, CGA, AUA, CUA, GGA, and CCC. If the gene of interest is wild-type, it is advisable to first analyze the frequency of rare codons. For genes with a low frequency of rare codons, using a Rosetta strain can enhance expression. However, if the gene contains a high frequency of rare codons, codon optimization followed by gene synthesis is the best approach. Codon optimization involves selecting codons that are more frequently used in E. coli (typically those with a usage frequency >20%). This process can improve the stability of mRNA secondary structures and prevent translation delays, premature termination, frameshifts, and amino acid mismatches caused by insufficient tRNA pools (Gustafsson et al., 2004).
3. Selection of Appropriate Expression Vectors
Even with an optimized gene sequence, achieving soluble protein expression requires the right expression vector. The pET28 series is widely applicable for various proteins. The pET22b vector can direct the secretion of target proteins into the periplasmic space, which facilitates disulfide bond formation and proper protein folding, thereby enhancing solubility. Fusion systems such as pGEX4T, pET32, and pET43.1 can also improve solubility by incorporating fusion tags that promote solubility. Additionally, the pCold and pBAD series of vectors are excellent choices for optimizing protein expression conditions (Rosano and Ceccarelli, 2014).
4. Choice of Suitable Expression Strains
After selecting the appropriate expression vector, the next step is to choose the right E. coli strain for protein expression. The BL21 strain is commonly used for routine protein expression. The T7E strain can enhance protein expression levels. The Origami strain is designed to promote disulfide bond formation within the cytoplasm. The Arctic strain allows for induction at 12°C, which can improve protein solubility and activity (Takai et al., 2001).
5. Optimization of Expression Conditions
Achieving soluble protein expression requires a comprehensive approach that considers all aspects of the expression system. This includes testing the compatibility of expression vectors with different E. coli strains and optimizing induction conditions, such as selecting the appropriate growth medium, adjusting induction temperature, and fine-tuning inducer concentrations. These optimizations can significantly reduce the formation of inclusion bodies (Baneyx and Mujacic, 2004).
Conclusion
While the formation of inclusion bodies is a common challenge in E. coli protein expression, it is not always a disadvantage. Inclusion bodies often yield high protein expression levels and are easier to purify, making them a suitable choice for applications such as antibody production for Western blotting. However, for obtaining biologically active proteins, the strategies outlined above can significantly enhance protein solubility and functionality. By carefully considering the removal of signal peptides, codon optimization, selection of appropriate vectors and strains, and optimization of expression conditions, researchers can effectively overcome the challenges posed by inclusion bodies and achieve successful protein expression in E. coli systems.
References
1. Baneyx, F. (1999). Recombinant protein expression in Escherichia coli. Current Opinion in Biotechnology, 10(5), 411-421.
2. Baneyx, F., & Mujacic, M. (2004). Recombinant protein folding and misfolding in Escherichia coli. Nature Biotechnology, 22(11), 1399-1408.
3. Gustafsson, C., Govindarajan, S., & Minshull, J. (2004). Codon bias and heterologous protein expression. Trends in Biotechnology, 22(7), 346-353.
4. Rosano, G. L., & Ceccarelli, E. A. (2014). Recombinant protein expression in Escherichia coli: advances and challenges. Frontiers in Microbiology, 5, 172.
5. Takai, K., Nakajima, R., & Tanokura, M. (2001). Expression of soluble and functional human liver fatty acid-binding protein in Escherichia coli using the cold-shock expression system. Protein Expression and Purification, 22(2), 245-251.

