How to use AlphaFold3 to predict protein structures
Molecular modeling serves as an indispensable technique in biological research, allowing scientists to delve into the architecture and functionality of biomolecules. Among the various tools available for this purpose, Alphafold 3 stands out as a cutting-edge application. This advanced deep learning model is capable of predicting protein structures with remarkable precision. Here is the information on how to use AlphaFold3.
I. Website and Login
The server URL for AlphaFold3 is:
https://golgi.sandbox.google.com
II. Complex Structure Prediction
Protein-Metal Ions
The supported metal ions include Mg, Zn, Cl, Ca, Na, Mn, K, Fe, Cu, Co, these ten metal ions. Take 4R59 as an example, which combines with the metal ion zinc: Enter the protein sequence, then click Add Entity, and select Ion
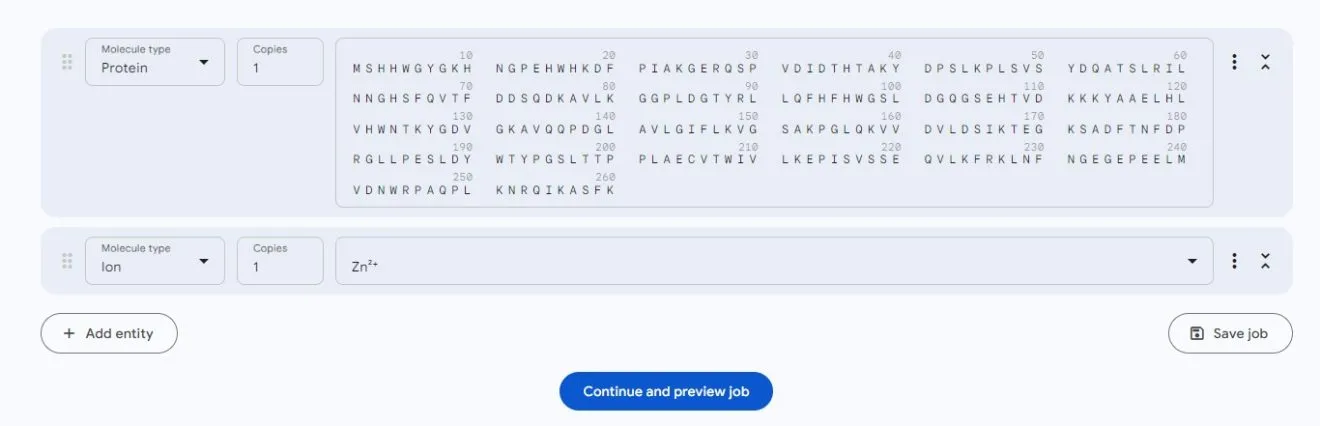
Click continue and preview, and after a while, you will get the results:
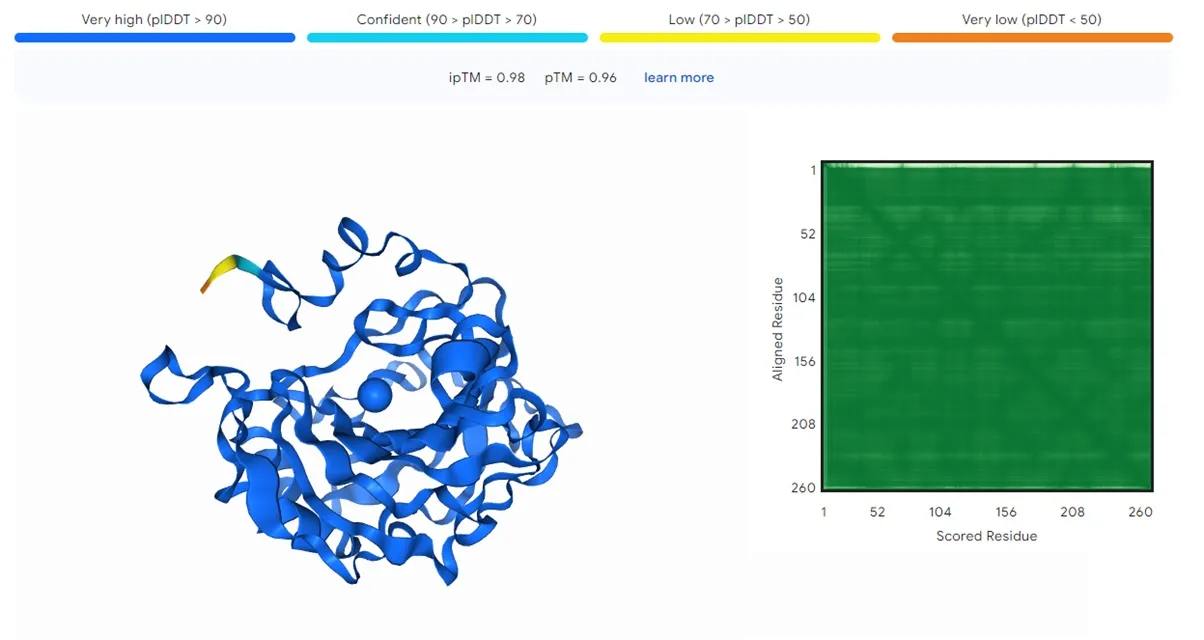
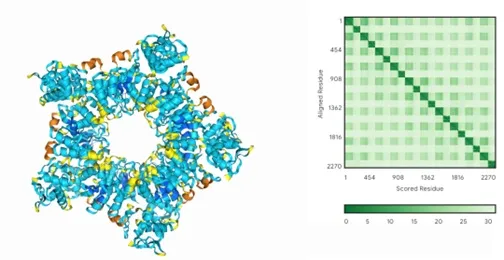
The color of the markings above indicates the confidence of the predicted structure, with blue being the best and orange being the worst. The error matrix on the right, the greener, the better the prediction.
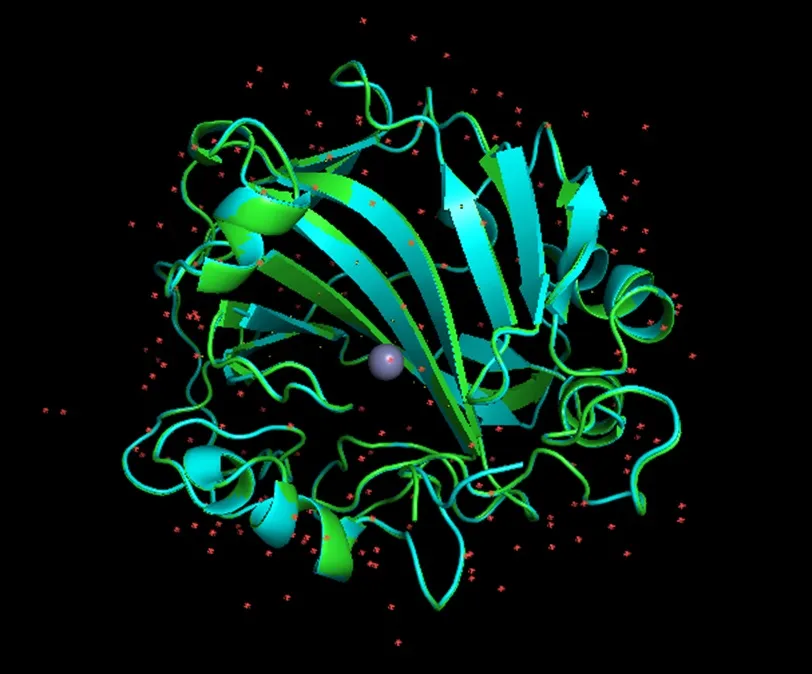
When aligned the predicted structure with the crystal structure, and the RMSD obtained is 0.113, which is a very good result, and the metal sites basically coincide.
Protein-Small Molecules
Unlike metal ions, select Ligand, the server only supports nineteen ligands, which are ADP, ATP, AMP, GTP, GDP, FAD, NAD, NAP, NDP, HEM, HEC, PLM, OLA, MYR, CIT, CLA, CHL, BCL, BCB.
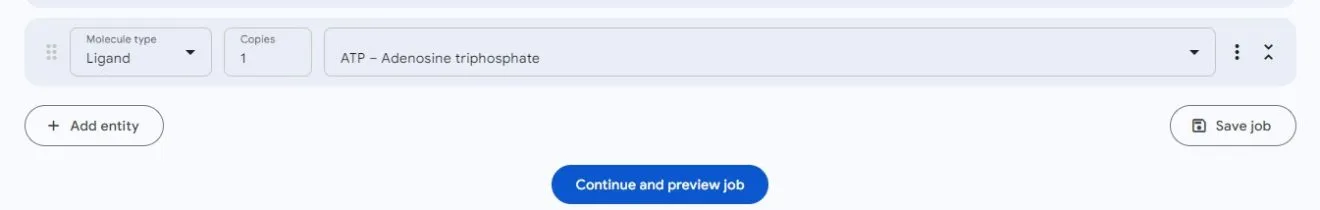
Protein-Nucleic Acids
DNA supports sequence input:
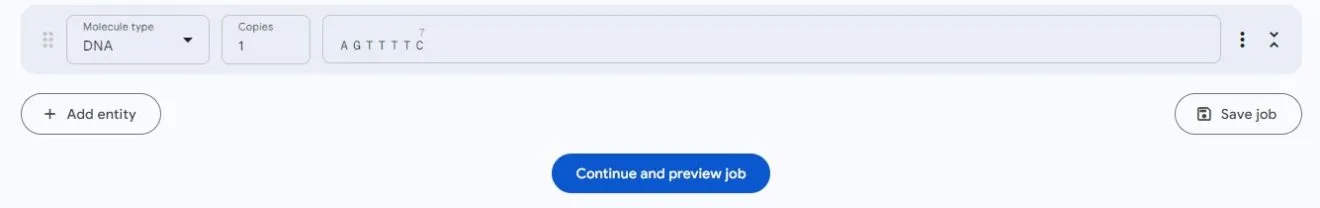
RNA also supports sequence input:
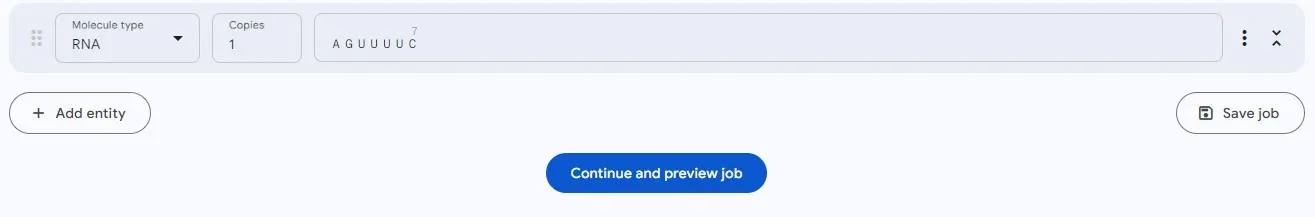
For Non-Standard Amino Acids and Nucleic Acids
For processing non-standard amino acids and nucleic acids, click on the red part:
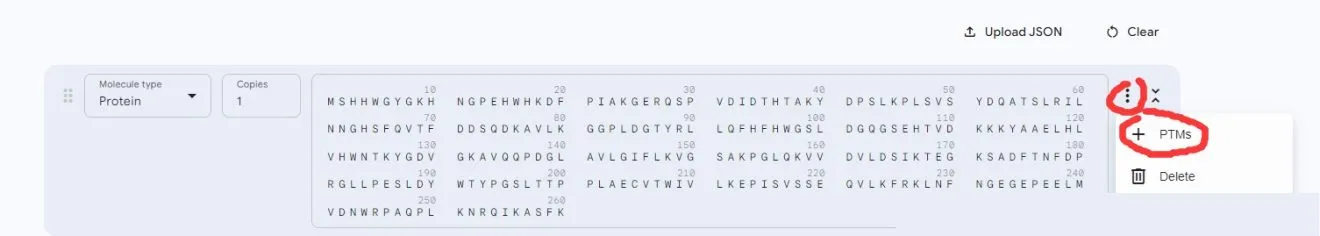
Then the following interface will appear, click on the amino acid to be modified:
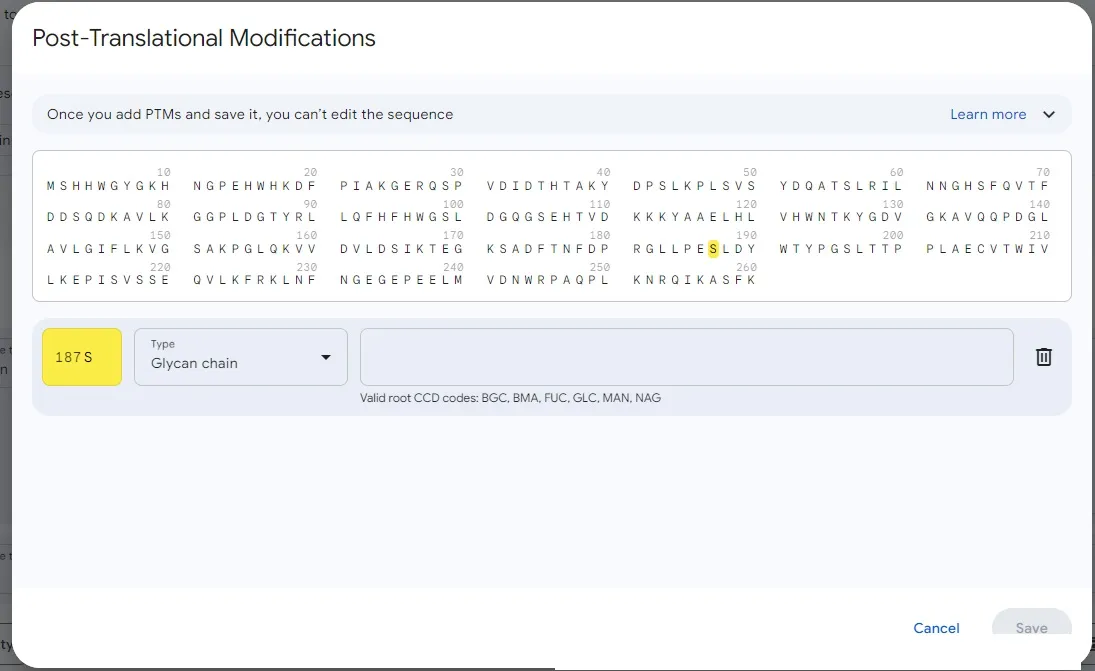
Then, for example, the S here can be changed to NAG, and then click save. DNA has Reverse and Modification operations, RNA only has Modification.
III. Parameters
Copies

For multimers, the structure is built by setting the copies here. For example, for a pentameric protein, setting the copies to 5 will result in:
Seed
The seed is an initial seed, and different settings can result in different structures. For example, the following sets the seed to random, 1, 2, 3, and 100:
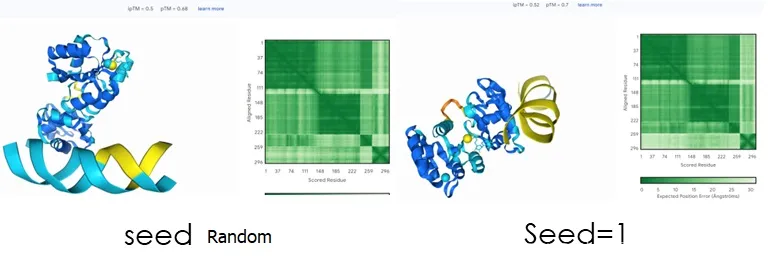
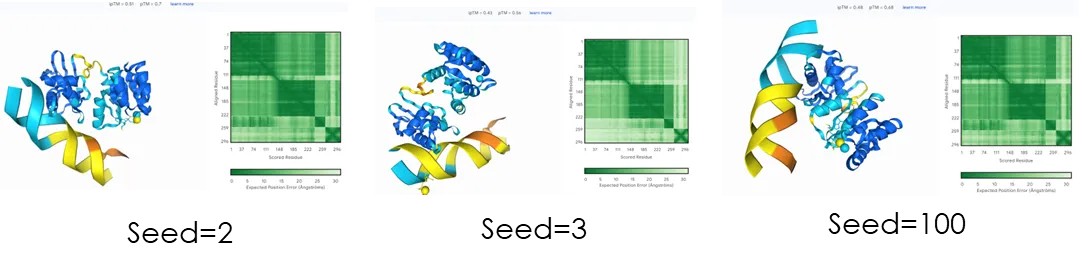
It can be seen that the obtained structural metal centers are different, and the position of nucleic acids also has a larger difference. However, the seed is not the larger the better. The results of 2 and 100 here are close, but 100 will take more time. Therefore, the specific settings should be based on the characteristics of the required structure.
