Summary
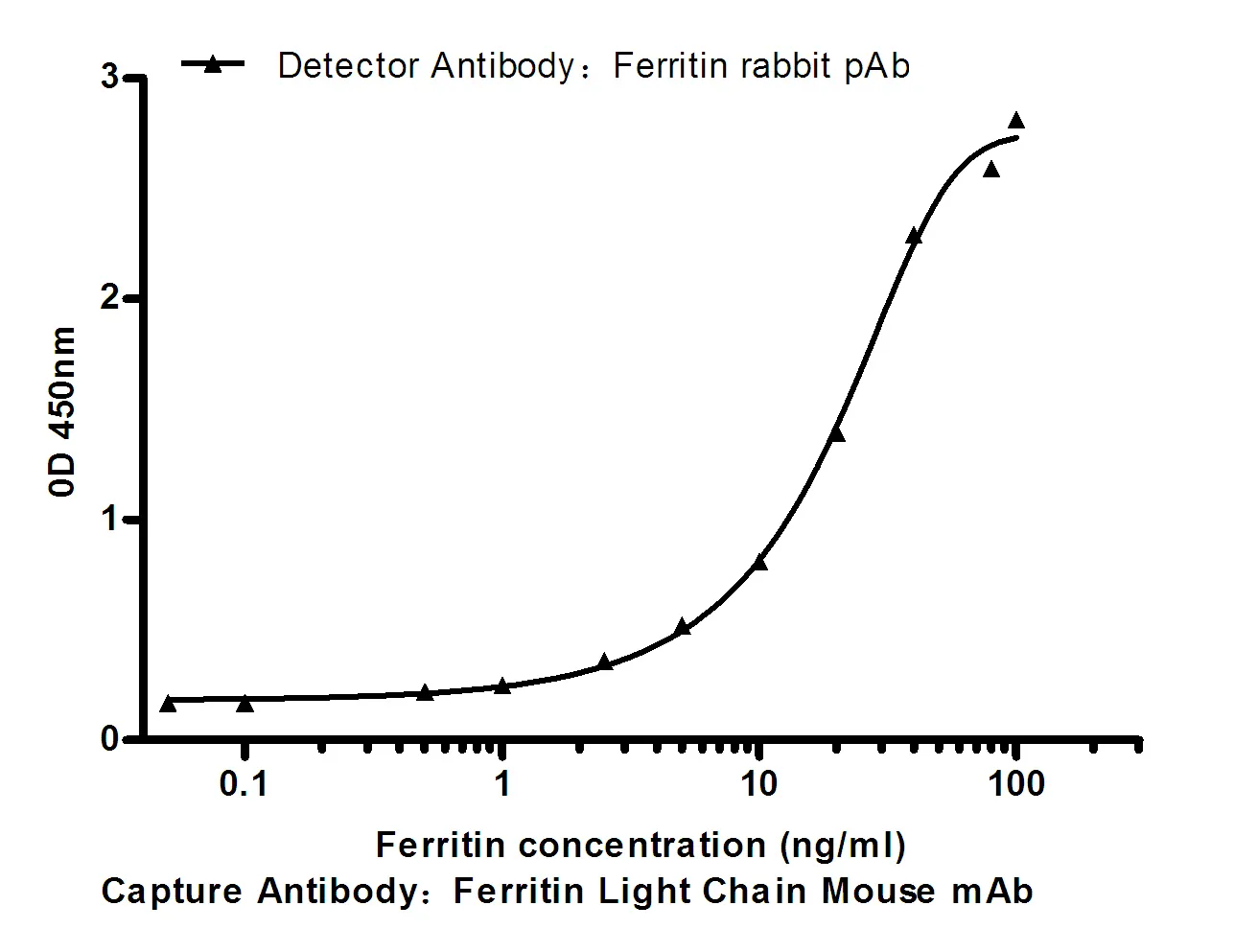
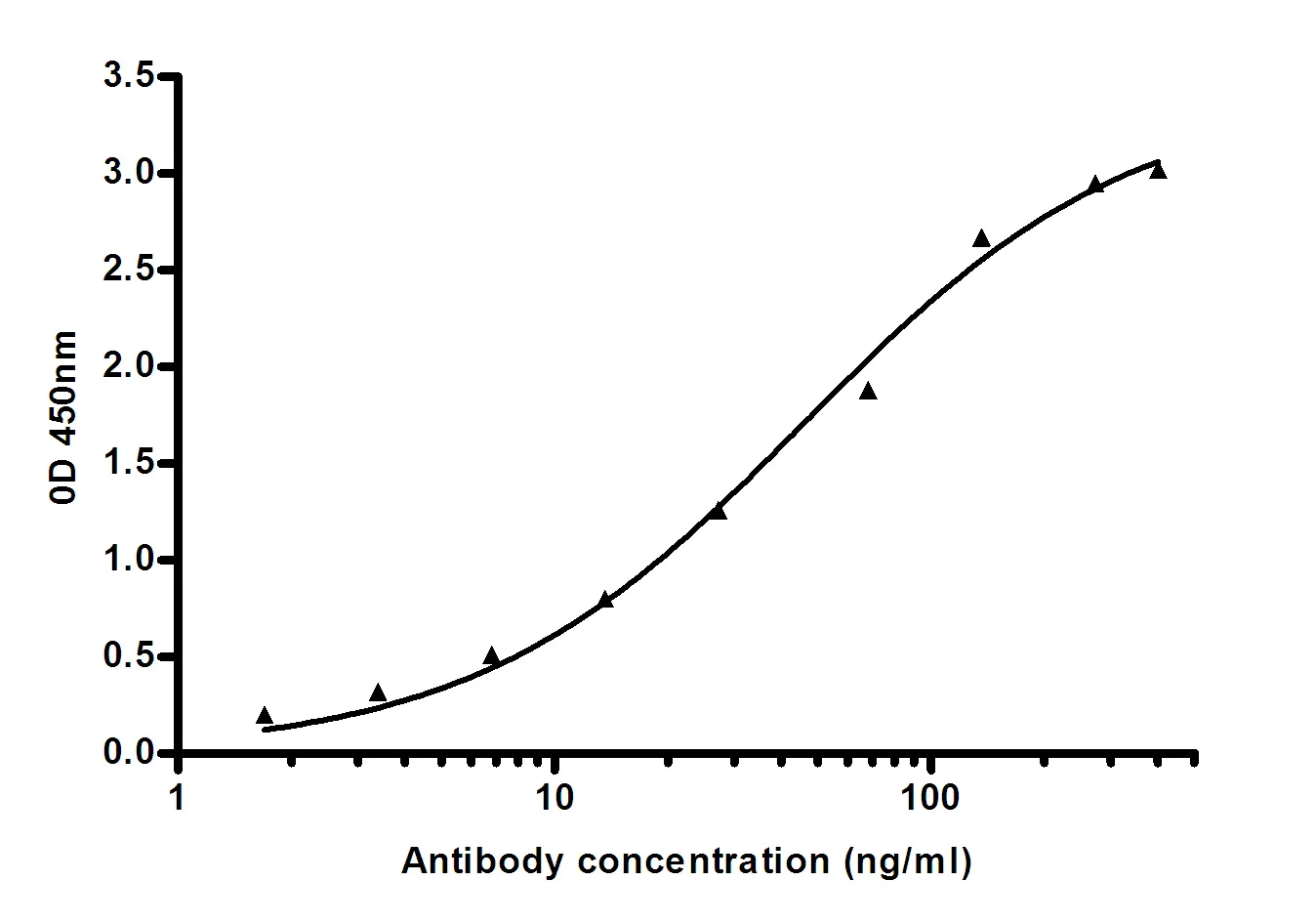
Performance
Immunogen
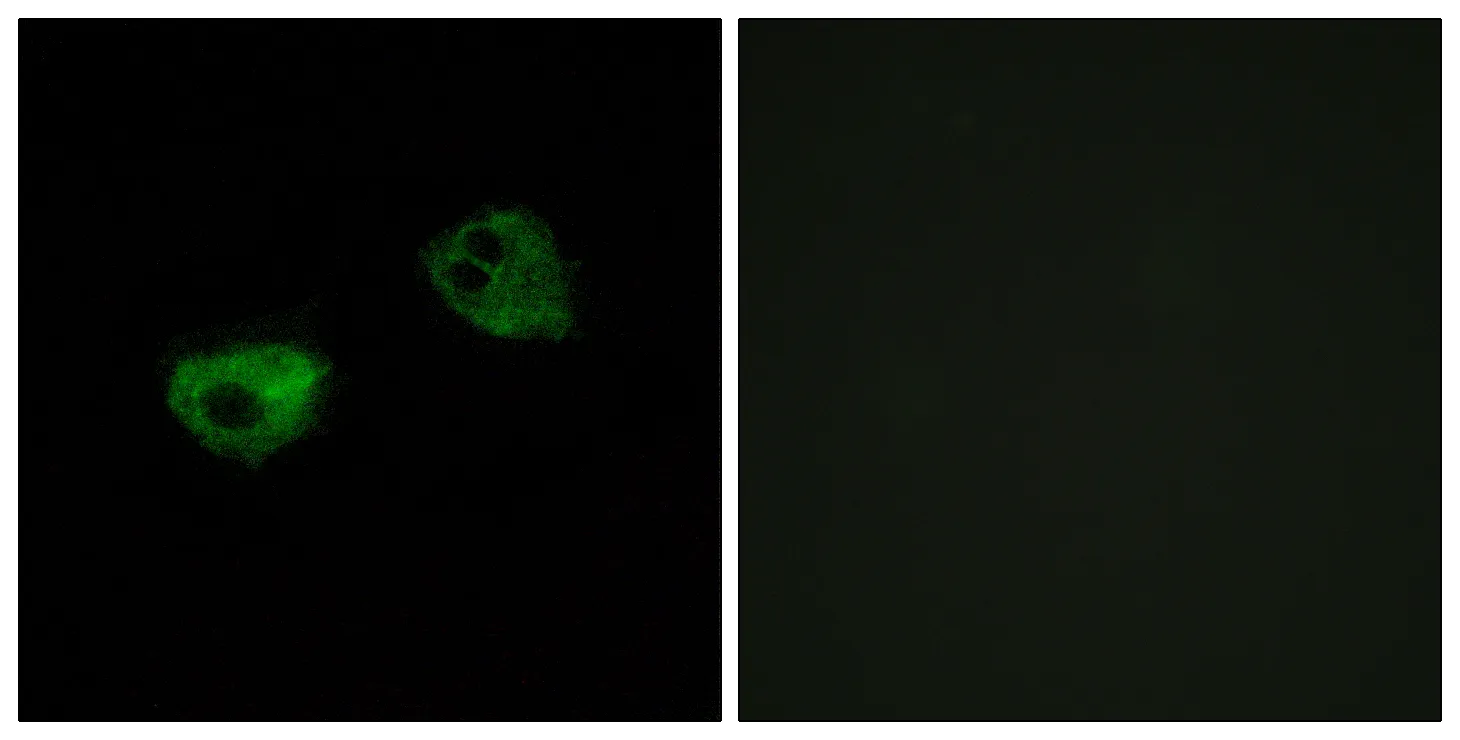
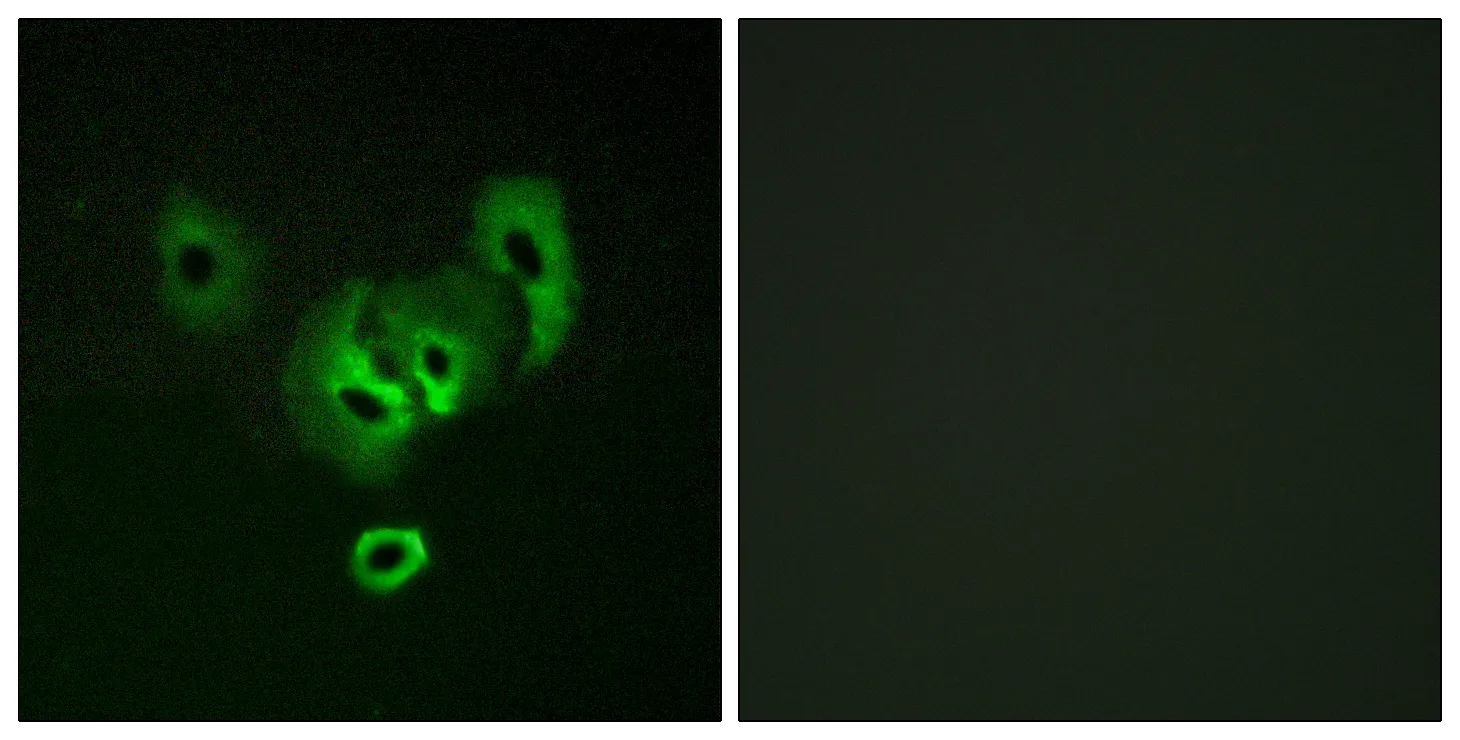
Application
Background
function:May have a role in checkpoint signaling during mitosis (By similarity). Enhances TP53-mediated transcriptional activation. Plays a role in the response to DNA damage.,PTM:Asymmetrically dimethylated on Arg residues by PRMT1. Methylation is required for DNA binding.,PTM:Phosphorylated at basal level in the absence of DNA damage. Hyper-phosphorylated in an ATM-dependent manner in response to DNA damage induced by ionizing radiation. Hyper-phosphorylated in an ATR-dependent manner in response to DNA damage induced by UV irradiation.,similarity:Contains 2 BRCT domains.,subcellular location:Associated with kinetochores. Both nuclear and cytoplasmic in some cells. Recruited to sites of DNA damage, such as double stand breaks. Methylation of histone H4 at 'Lys-20' is required for efficient localization to double strand breaks.,subunit:Interacts with IFI202A (By similarity). Binds to the central domain of TP53/p53. May form homo-oligomers. Interacts with DCLRE1C. Interacts with histone H2AFX and this requires phosphorylation of H2AFX on 'Ser-139'. Interacts with histone H4 that has been dimethylated at 'Lys-20'. Has low affinity for histone H4 containing monomethylated 'Lys-20'. Does not bind histone H4 containing unmethylated or trimethylated 'Lys-20'. Has low affinity for histone H3 that has been dimethylated on 'Lys-79'. Has very low affinity for histone H3 that has been monomethylated on 'Lys-79' (in vitro). Does not bind unmethylated histone H3.,function:May have a role in checkpoint signaling during mitosis (By similarity). Enhances TP53-mediated transcriptional activation. Plays a role in the response to DNA damage.,PTM:Asymmetrically dimethylated on Arg residues by PRMT1. Methylation is required for DNA binding.,PTM:Phosphorylated at basal level in the absence of DNA damage. Hyper-phosphorylated in an ATM-dependent manner in response to DNA damage induced by ionizing radiation. Hyper-phosphorylated in an ATR-dependent manner in response to DNA damage induced by UV irradiation.,similarity:Contains 2 BRCT domains.,subcellular location:Associated with kinetochores. Both nuclear and cytoplasmic in some cells. Recruited to sites of DNA damage, such as double stand breaks. Methylation of histone H4 at 'Lys-20' is required for efficient localization to double strand breaks.,subunit:Interacts with IFI202A (By similarity). Binds to the central domain of TP53/p53. May form homo-oligomers. Interacts with DCLRE1C. Interacts with histone H2AFX and this requires phosphorylation of H2AFX on 'Ser-139'. Interacts with histone H4 that has been dimethylated at 'Lys-20'. Has low affinity for histone H4 containing monomethylated 'Lys-20'. Does not bind histone H4 containing unmethylated or trimethylated 'Lys-20'. Has low affinity for histone H3 that has been dimethylated on 'Lys-79'. Has very low affinity for histone H3 that has been monomethylated on 'Lys-79' (in vitro). Does not bind unmethylated histone H3.,
Research Area