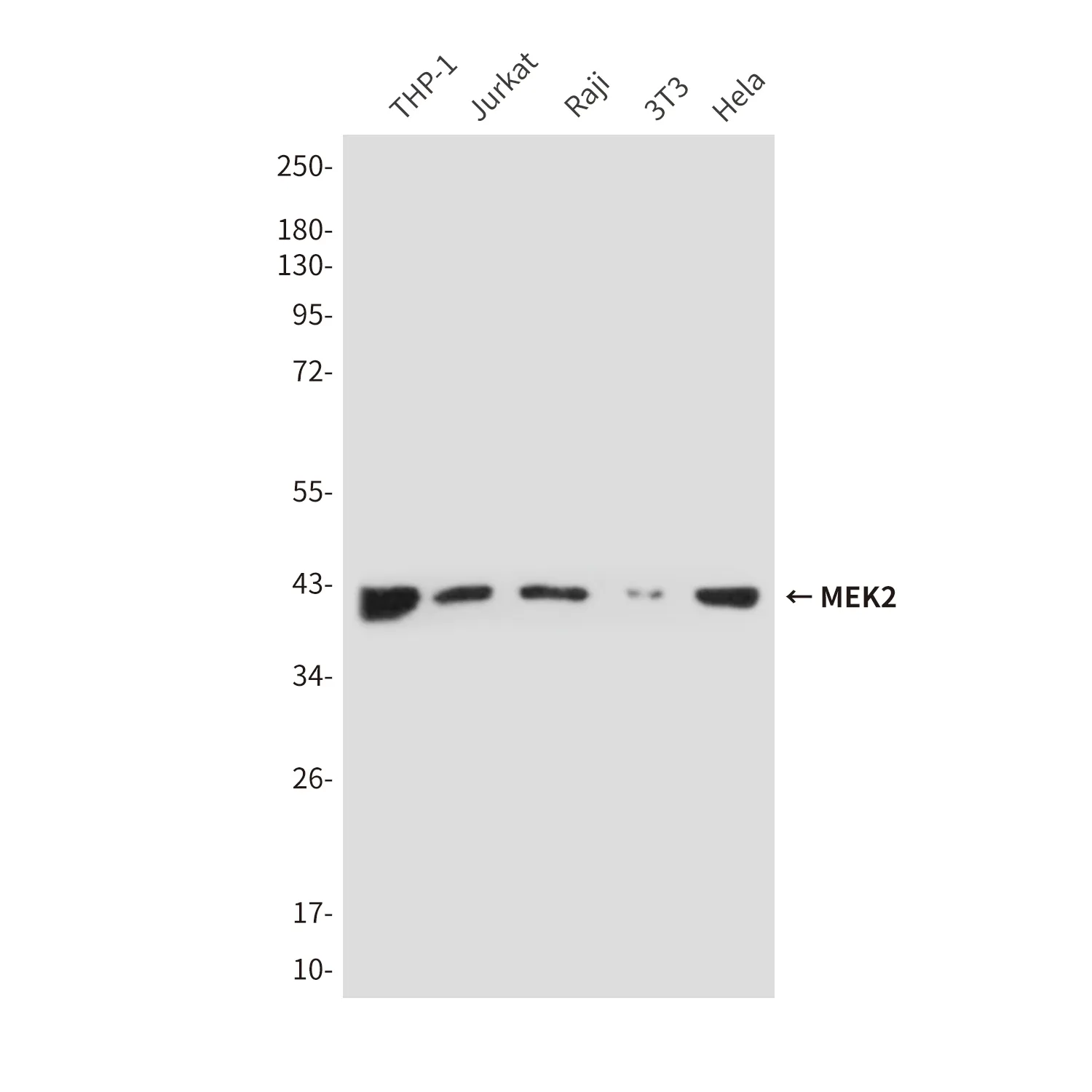
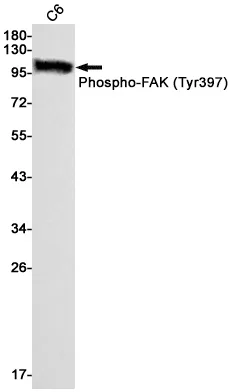
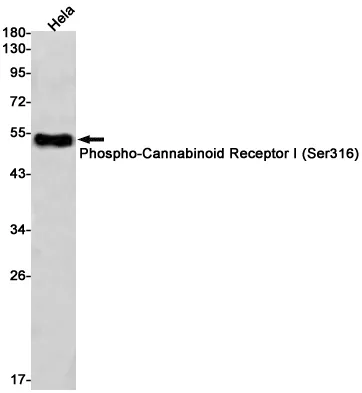
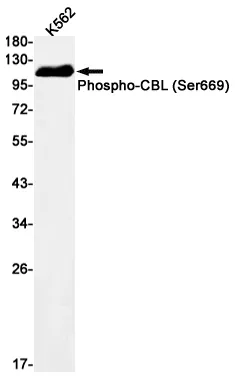
Summary
Performance
Immunogen
Application
Background
Has both an apurinic and/or apyrimidinic endonuclease activity and a DNA N-glycosylase activity. Incises damaged DNA at cytosines, thymines and guanines. Acts on a damaged strand, 5' from the damaged site. Required for the repair of both oxidative DNA damage and spontaneous mutagenic lesions. Bifunctional DNA N-glycosylase with associated apurinic/apyrimidinic (AP) lyase function that catalyzes the first step in base excision repair (BER), the primary repair pathway for the repair of oxidative DNA damage (PubMed:9927729). The DNA N-glycosylase activity releases the damaged DNA base from DNA by cleaving the N- glycosidic bond, leaving an AP site. The AP-lyase activity cleaves the phosphodiester bond 3' to the AP site by a beta-elimination. Primarily recognizes and repairs oxidative base damage of pyrimidines. Has also 8-oxo-7,8-dihydroguanine (8-oxoG) DNA glycosylase activity. Acts preferentially on DNA damage opposite guanine residues in DNA. Is able to process lesions in nucleosomes without requiring or inducing nucleosome disruption.
Research Area