Summary
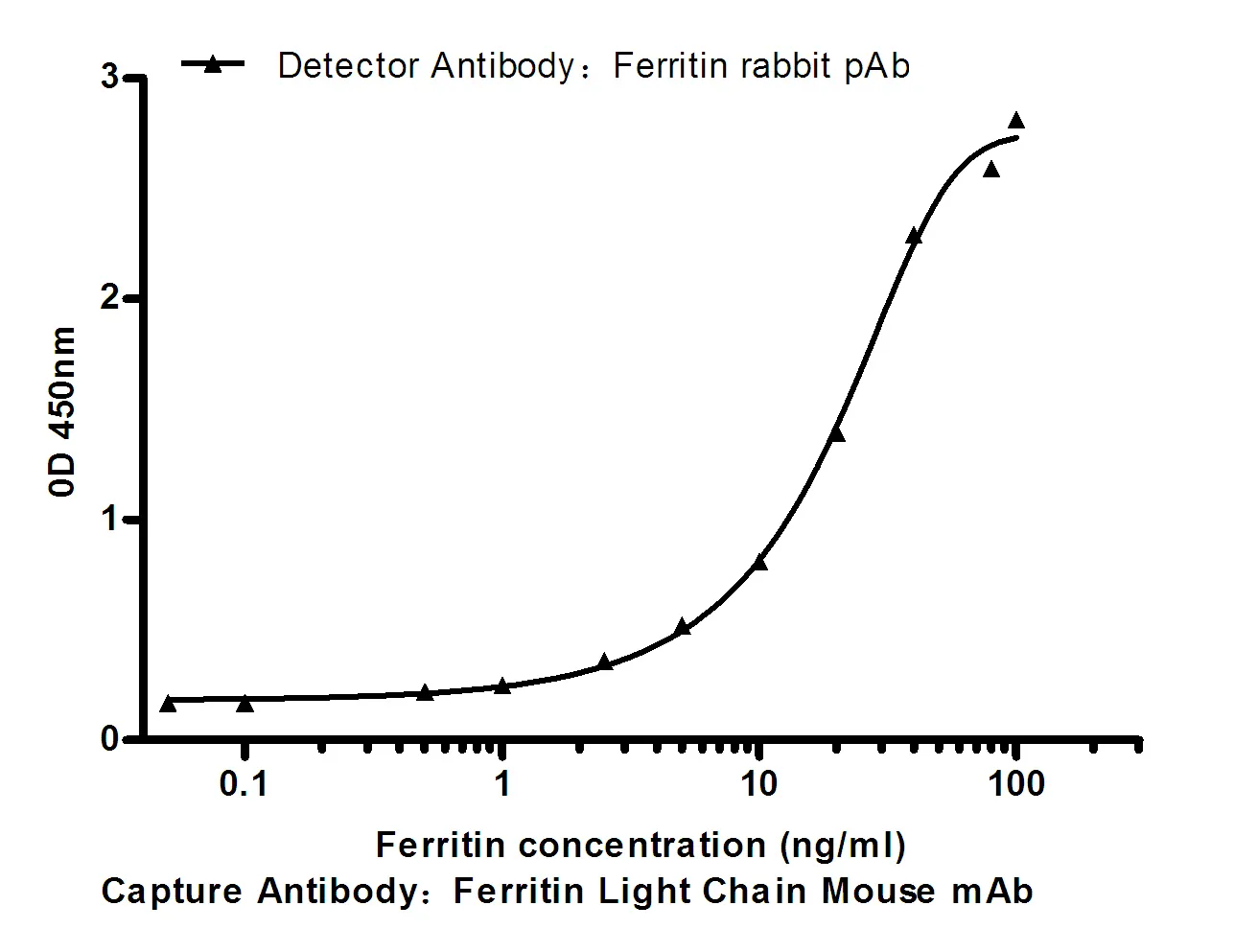
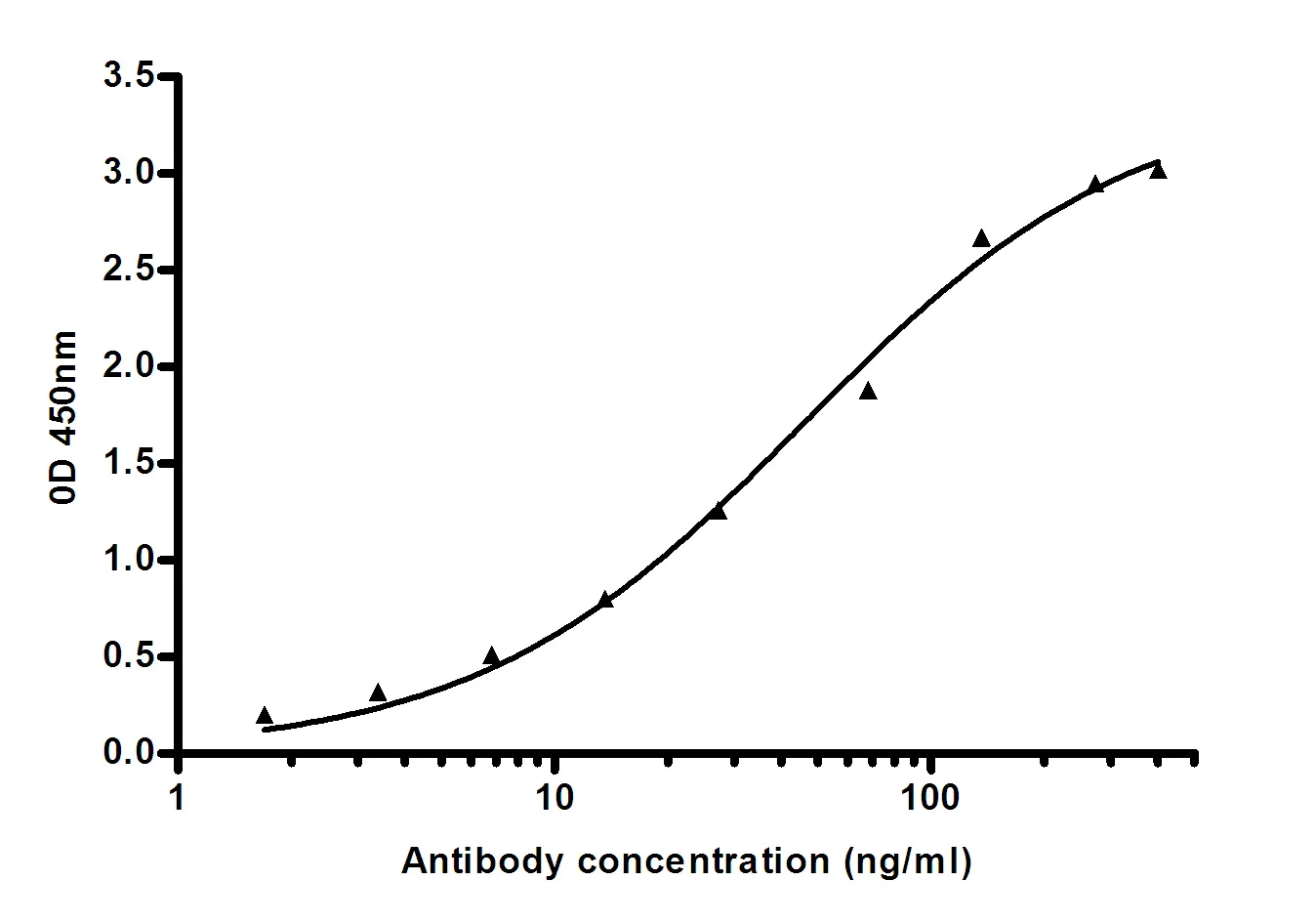
Performance
Immunogen
Application
Background
E1A binding protein p300(EP300) Homo sapiens This gene encodes the adenovirus E1A-associated cellular p300 transcriptional co-activator protein. It functions as histone acetyltransferase that regulates transcription via chromatin remodeling and is important in the processes of cell proliferation and differentiation. It mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. This gene has also been identified as a co-activator of HIF1A (hypoxia-inducible factor 1 alpha), and thus plays a role in the stimulation of hypoxia-induced genes such as VEGF. Defects in this gene are a cause of Rubinstein-Taybi syndrome and may also play a role in epithelial cancer. [provided by RefSeq, Jul 2008],catalytic activity:Acetyl-CoA + histone = CoA + acetylhistone.,disease:Chromosomal aberrations involving EP300 may be a cause of acute myeloid leukemias. Translocation t(8;22)(p11;q13) with MYST3.,disease:Defects in EP300 are a cause of Rubinstein-Taybi syndrome (RSTS) [MIM:180849]. RSTS is an autosomal dominant disorder characterized by craniofacial abnormalities, broad thumbs, broad big toes, mental retardation and a propensity for development of malignancies.,disease:Defects in EP300 may play a role in epithelial cancer.,function:Functions as histone acetyltransferase and regulates transcription via chromatin remodeling. Acetylates all four core histones in nucleosomes. Histone acetylation gives an epigenetic tag for transcriptional activation. Binds to and may be involved in the transforming capacity of the adenovirus E1A protein. Mediates cAMP-gene regulation by binding specifically to phosphorylated CREB protein. In case of HIV-1 infection, it is recruited by the viral protein Tat. Regulates Tat's transactivating activity and may help inducing chromatin remodeling of proviral genes.,online information:P300/CBP entry,PTM:Acetylated on Lys at up to 17 positions by intermolecular autocatalysis.,PTM:Citrullinated at Arg-2142 by PADI4, which impairs methylation by CARM1 and promotes interaction with NCOA2/GRIP1.,PTM:Methylated at Arg-580 and Arg-604 in the KIX domain by CARM1, which blocks association with CREB, inhibits CREB signaling and activates apoptotic response. Also methylated at Arg-2142 by CARM1, which impairs interaction with NCOA2/GRIP1.,PTM:Phosphorylated.,similarity:Contains 1 bromo domain.,similarity:Contains 1 KIX domain.,similarity:Contains 1 ZZ-type zinc finger.,similarity:Contains 2 TAZ-type zinc fingers.,subunit:Interacts with phosphorylated CREB1 (By similarity). Interacts with DTX1, EID1, ELF3, FEN1, LEF1, NCOA1, NCOA6, NR3C1, PCAF, PELP1, PRDM6, SPIB, SRY, TCF7L2, TP53, SRCAP, TTC5, JMY and TRERF1. The TAZ-type 1 domain interacts with HIF1A. Probably part of a complex with HIF1A and CREBBP. Part of a complex containing CARM1 and NCOA2/GRIP1. Interacts with ING4 and this interaction may be indirect. Interacts with ING5. Interacts with the C-terminal region of CITED4. Interacts with HTLV-1 Tax and p30II. Interacts with and acetylates HIV-1 Tat.,
Research Area
Cell_Cycle_G1S;Cell_Cycle_G2M_DNA; WNT;WNT-T CELL;β-Catenin; Protein_Acetylation