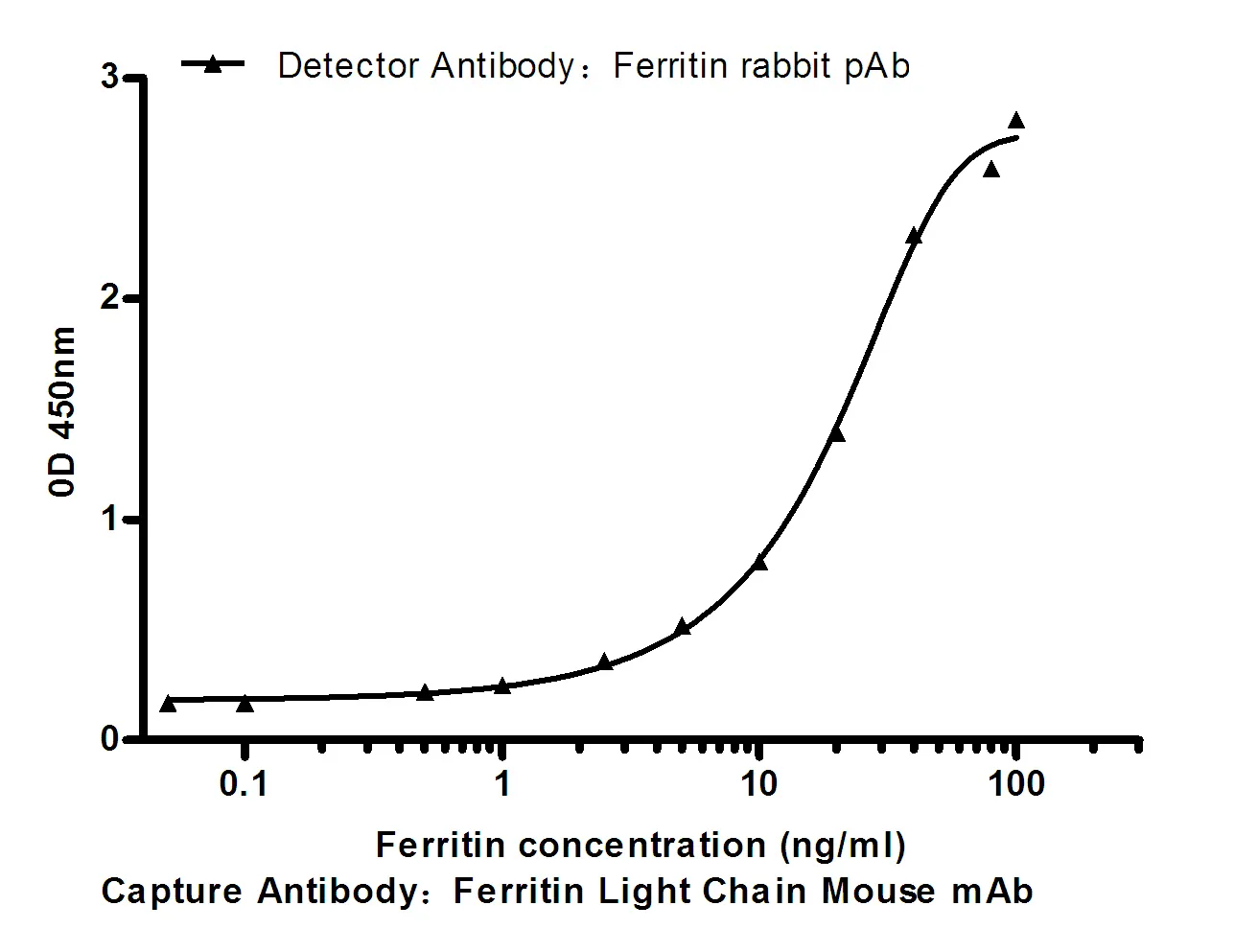
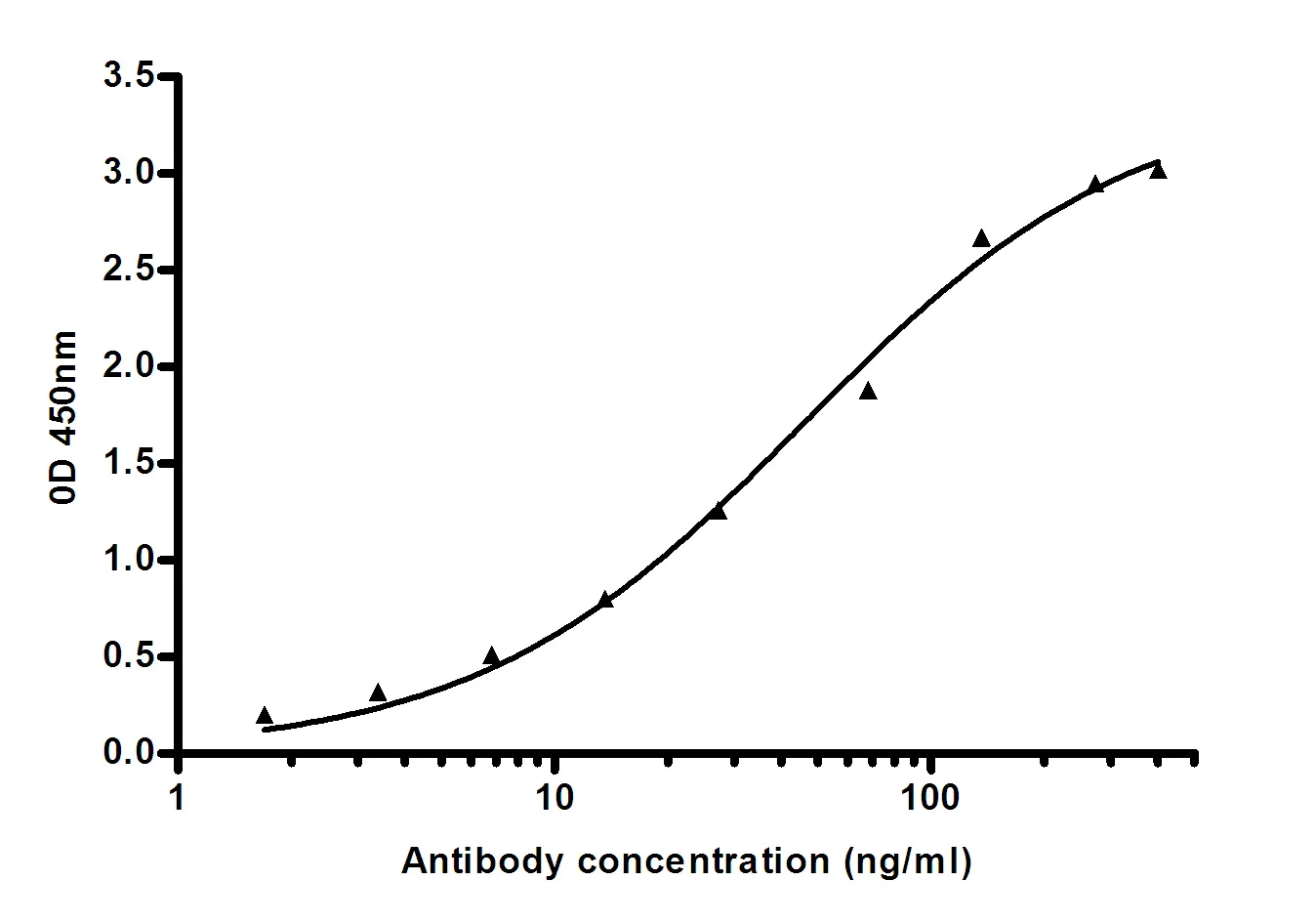
Summary
Performance
Immunogen
Application
Background
The protein encoded by this gene belongs to the PARP family. These enzymes modify nuclear proteins by poly-ADP-ribosylation, which is required for DNA repair, regulation of apoptosis, and maintenance of genomic stability. This gene encodes the poly(ADP-ribosyl)transferase 3, which is preferentially localized to the daughter centriole throughout the cell cycle. Alternatively spliced transcript variants encoding different isoforms have been identified. [provided by RefSeq, Jul 2008],catalytic activity:NAD(+) + (ADP-D-ribosyl)(n)-acceptor = nicotinamide + (ADP-D-ribosyl)(n+1)-acceptor.,domain:According to PubMed:10329013 the N-terminal domain (54 amino acids) of isoform 2 is responsible for its centrosomal localization. The C-terminal region contains the catalytic domain.,function:Involved in the base excision repair (BER) pathway, by catalyzing the poly(ADP-ribosyl)ation of a limited number of acceptor proteins involved in chromatin architecture and in DNA metabolism. This modification follows DNA damages and appears as an obligatory step in a detection/signaling pathway leading to the reparation of DNA strand breaks. May link the DNA damage surveillance network to the mitotic fidelity checkpoint. Negatively influences the G1/S cell cycle progression without interfering with centrosome duplication. Binds DNA. May be involved in the regulation of PRC2 and PRC3 complex-dependent gene silencing.,PTM:Auto-poly(ADP)-ribosylation.,similarity:Contains 1 PARP alpha-helical domain.,similarity:Contains 1 PARP catalytic domain.,subcellular location:Core component of the centrosome. Preferentially localized to the daughter centriole throughout the cell cycle. According PubMed:16924674 is almost exclusively localized in the nucleus and appears in numerous small foci and a small number of larger foci whereas a centrosomal location has not been detected.,subunit:Interacts with PRKDC and PARP1. Interacts with XRCC5; the interaction is dependent on nucleic acids. Interacts with XRCC6; the interaction is dependent on nucleic acids. Interacts with EZH2, HDAC1, HDAC2, SUZ12, YY1, LIG3 and LIG4.,tissue specificity:Widely expressed; the highest levels are in the kidney, skeletal muscle, liver, heart and spleen; also detected in pancreas, lung, placenta, brain, leukocytes, colon, small intestine, ovary, testis, prostate and thymus.,
Research Area
Base excision repair;